SCORE-SeqTDS
SCORE-SeqTDS: Score Tests for Sequencing Studies with Trait-Dependent Sampling
General information
SCORE-SeqTDS is a command-line program written in the C language to implement the methods described in Lin et al. (2013) for analyzing primary and secondary quantitative traits under trait-dependent sampling. The primary trait is the trait that is used to select subjects for sequencing, and all other traits are treated as secondary. Each quantitative trait is related to a genetic variable and possibly covariates through a linear regression model. Both the maximum likelihood estimation (MLE) and standard least-squares (LS) methods are available. The MLE method properly accounts for trait-dependent sampling whereas the LS method does not. The LS method is the ideal choice for random sampling and is approximately correct for analyzing secondary quantitative traits in case-control or case-only studies with rare diseases. SCORE-SeqTDS performs the LS analysis on secondary quantitative traits for random sampling, case-control and case-only sampling. For random sampling, all traits are treated as secondary (because the sampling does not depend on any particular trait.) The sampling scheme is specified through the option -sampling. A table in the OPTIONS section below summarizes the available analysis options for different sampling schemes.
The genetic variable pertains to the genotype in the single-variant analysis and to the burden score in the rare-variant analysis. The burden score may be determined externally (default) or internally. Use the option -gfile to specify the file that contains the external genetic variables. Otherwise, use the options -gfile and -mfile to specify the genotype file and the mapping file, respectively, for the internal creation of the burden scores. Use the option -test to request one of the six tests (T1, T5, MB, VT, SKAT, and customized test) under the additive genetic model (default). The T1, T5, VT, and SKAT tests under the dominant (recessive) genetic model can be obtained by using the option -dominant (-recessive). There are options for the minor allele frequency (MAF) upper bound, the minor allele count (MAC) lower bound and the call rate (CR) lower bound. Under the additive or dominant (recessive) model, MAC is defined as the number of subjects with at least one (two) observed mutation. A genetic variable is excluded from analysis if its MAC is smaller than the MAC lower bound or its CR is smaller than the CR lower bound. When the burden score is created internally, a variant is deleted if its MAF is greater than the MAF upper bound or its CR is smaller than the CR lower bound. By default, the MAF upper bound is 0.05, the MAC lower bound is 1 and the CR lower bound is 0. The MAFs may be determined internally (i.e., calculated from the genotype file) or externally (i.e., input in the mapping file). The MAF thresholds in the VT test can be determined internally (i.e., based on the unique MAFs among the aggregating variants) or externally (i.e., input in the mapping file).
If the quantitative trait of interest is measured in multiple studies, either as primary or secondary trait, then SCORE-SeqTDS can be used to analyze the trait of interest for one study at a time, and the results can be combined through meta-analysis. For T1, T5 and MB, SCORE-SeqTDS outputs the score statistics and their variance estimates. For the VT, SKAT and customized tests, SCORE-SeqTDS outputs the score vector and the information matrix. The results from multiple studies can be combined through the accompanying program MASS.
SYNOPSIS
SCORE-SeqTDS [-sampling sampling] [-method method] [-nsec nsec] [-pfile_seq phenofile.seq] [-cov cov] [-pfile_nonseq phenofile.nonseq] [-rounding NMAX] [-gfile genofile] [-mfile mapfile] [-wfile wfile] [-test test] [-dominant ] [-recessive ][-ofile outfile] [-msglog msglog] [-MAF MAF_UB] [-MAC MAC_LB] [-CR CR_LB] [-log ] [-INT ] [-R-INT SD_TYPE]
OPTIONS
| Option | Parameter | Default | Description |
|---|---|---|---|
| -sampling | {sampling} | TDS | Specify the sampling design of the study. There are three options: “TDS” (trait dependent sampling), “CC” (case-control sampling), and “random” (case-only or random sampling). The default is “TDS”. |
| -method | {method} | MLE | Specify the method to be used. There are two options: MLE and LS. For trait-dependent sampling, the default is MLE. For random sampling, case-control and case-only sampling, this option is invalid because only the LS method is applicable. |
| -nsec | {nsec} | 0 for trait-dependent sampling;
1 for random sampling, case-control and case-only sampling |
Specify the number of secondary traits (nsec) to be analyzed. nsec = 0 means to analyze the primary trait only. For random-sampling, case-control and case-only sampling, nsec should be set to an integer ≥ 1 since only secondary traits are analyzed. |
| -pfile_seq | {phenofile.seq} | pheno_seq.txt |
Specify the file that contains the trait(s) and covariates (if any) for the sequenced subjects. |
| -cov | {cov} | No | Specify the file for selecting covariates. |
| -pfile_nonseq | {phenofile.nonseq} | pheno_nonseq.txt |
Specify the file that contains the primary trait for the non-sequenced subjects. |
| -rounding | {NMAX} | 500 |
Round the values of the primary trait such that the number of unique values is at most NMAX, which is any positive integer. It is suggested that NMAX be smaller than 500 to speed up computation. This option is only valid under TDS. |
| -gfile | {genofile} | geno.txt |
Specify the genotype file. The file content is described in the table of the INPUT FILES section. |
| -mfile | {mapfile} | mapping.txt |
Specify the gene-SNP mapping file. |
| -wfile | {wfile} | No | Specify the file that contains the customized weight matrix. |
| -test | {test} | Single variant test | Specify the rare-variant test to be performed. There are six options: T1, T5, MB, VT, SKAT, and customized. Refer to the Options section of the software SCORE-Seq for a detailed description of these tests. |
| -dominant | No | Use the dominant genetic model for the T1, T5, VT, and SKAT tests. | |
| -recessive | No | Use the recessive genetic model for the T1, T5, VT, and SKAT tests. | |
| -ofile | {outfile} | output.txt |
Specify the output file. |
| -msglog | {msglog} | log |
Specify the prefix for the names of log files. |
| -MAF | {MAF_UB} | 0.05 | Specify the MAF upper bound, which is any number between 0 and 1. |
| -MAC | {MAC_LB} | 1 | Specify the MAC lower bound, which is any integer. |
| -CR | {CR_LB} | 0 | Specify the call rate lower bound, which is any number between 0 and 1. |
| -log | No | Apply the natural logarithm to the primary trait. | |
| -INT | No | Apply inverse normal transformation (INT) to the trait of interest. | |
| -R-INT | {SD_TYPE} | 1 | Apply rescaled inverse normal transformation (R-INT) to the trait of interest using conventional standard deviation (if SD_TYPE=1) or the robust standard deviation (if SD_TYPE=2) described here http://en.wikipedia.org/wiki/Median_absolute_deviation. |
1. MLE method
The MLE method is based on the maximum likelihood estimation of an observed-data likelihood that reflects trait-dependent sampling and thus is valid and efficient. Let Y1 be the primary trait and Y2 be the secondary trait. Also, let G1 and G2 denote the corresponding genetic variables and Z1 and Z2 denote the corresponding sets of covariates. The joint distribution of Y1 and Y2 is formulated through a bivariate linear regression model in which (G1, Z1) and (G2, Z2) are treated as independent variables. Let β1 and β2 denote the regression coefficients of G1 for Y1 and G2 for Y2, respectively. Then the null hypothesis of no genetic effect on the primary trait corresponds to H0(1) : β1 = 0 while the null hypothesis of no genetic effect on the secondary trait corresponds to H0(2) : β2 = 0. An EM algorithm is used to maximize the observed-data likelihood and to calculate the score statistics for testing H0(1) : β1 = 0 and H0(2) : β2 = 0. The estimation results for β1 and β2 are also provided.
2. LS method
The LS method is based on the standard least-squares estimation. For studying the genetic effect on the primary trait, the LS method is applied to the linear regression model relating Y1 to (G1, Z1). For studying the genetic effect on the secondary trait, there are two versions of the LS method: LS-M is based on the linear regression of Y2 on (G2, Z2) and LS-C is based on the linear regression of Y2 on (Y1, G2, Z2). All hypothesis tests are based on the score statistics.
| Sampling design | MLE on the primary trait | MLE on secondary trait(s) | LS on the primary trait | LS on secondary trait(s) |
|---|---|---|---|---|
| Trait-dependent sampling | Option: (default) Output: MLE |
Option: -nsec nsec
Output: MLE |
Option: -method LS Output: LS |
Option: -method LS -nsec nsecOutput: LS-M, LS-C |
| Case-control sampling | N/A | N/A | N/A | Option: -sampling CC -nsec nsecOutput: LS-M, LS-C |
| Case-only sampling or random sampling |
N/A | N/A | N/A | Option: -sampling random -nsec nsecOutput: LS |
INPUT FILES
The input files consist of a phenotype file for the sequenced subjects, a phenotype file for the non-sequenced subjects, a genotype file for the sequenced subjects and a mapping file. In all input files, the field separator character should be tab-delimiter and missing values are denoted by “-999” or “NA”.
PHENOTYPE FILE FOR SEQUENCED SUBJECTS
The 1st row contains the header. The remaining rows represent sequenced subjects. Under trait dependent-sampling or case-control sampling, the 1st column pertains to the primary trait. For trait-dependent sampling, the primary trait is quantitative. For case-control sampling, the primary trait takes the value 0 or 1. Under case-only or random sampling, the 1st column is empty. If there are nsec secondary quantitative traits, then the 2nd to the (nsec + 1)th columns pertain to the secondary traits. The remaining columns are covariates if there are any. The order of subjects in the rows of this file should be identical to the order of subjects in the columns of the genotype file. No missing value is allowed on the primary trait and all variables should be numeric.
FILE FOR SELECTING COVARIATES
If the covariates are different among traits, a file with the selection indicators for covariates should be specified. This file contains one matrix with entries 0 or 1. The value of 1 indicates the covariate on the column is used by the trait on the row, the value of 0 indicates otherwise. The order of the traits on the rows and covariates on the columns in this file should be the same as the order in the phenotype file for sequenced subjects.
PHENOTYPE FILE FOR NON-SEQUENCED SUBJECTS
The first column contains the values of the primary trait for the non-sequenced subjects and the second column contains the frequencies of the trait values. If only the first column is provided, the trait values are all assumed to have frequencies of 1. No missing value is allowed. This file is not needed if the LS method is specified.
Note: To speed up computation, it is suggested that the values of the primary trait (1st column) in both phenotype files are rounded such that there is a total of at most 500 distinct values. If that is not the case, the program will automatically round the values to provide at most 500 unique values.
GENOTYPE FILE, MAPPING FILE, AND WEIGHT FILE
The table below shows the option setup, genotype file, mapping file, and weight file for different scenarios.
| Option | Genotype file | Mapping file and weight file | |
|---|---|---|---|
| Single-variant analysis | -gfile genofile | The 1st column contains the SNP ID. The remaining columns contain the genotypes. All genotype values are numeric. | N/A |
| Rare-variant analysis: burden scores provided by the user | -gfile genofile | The 1st column contains the gene ID. The remaining columns contain the burden scores. All scores are numeric. | N/A |
| Rare-variant analysis: T1, T5, MB, VT, or customized burden scores created internally | -test test
-gfile genofile -mfile mapfile |
The 1st column contains the SNP ID. The remaining columns contain the genotypes. The genotype takes the value 0, 1, or 2. | The mapping file contains at least two columns. The first column is the gene ID, and the second column is the SNP ID. Each row represents a unique gene-SNP pair. The third column contains the external MAFs if they are to be used in the analysis. The fourth column contains the external MAF threshold indicators, which indicate by the values 1 vs 0, whether the thresholds will be used in the VT test. For each gene, the total number of thresholds cannot exceed 20. To perform the customized test, a weight file should be specified. The first two columns of the weight file contain the same information as that of the mapping file and the remaining columns contain vectors of weights. No missing values are allowed in either the mapping file or the weight file. |
OUTPUT
OUTPUT FILE
- Tests with a single genetic variable (single-variant, T1, T5, or MB test)
The format is shown as follows: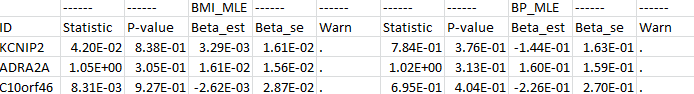 The 1st column contains the gene ID and the remaining columns under each main header <Trait_Method> contain the test statistic, the p-value, the genetic effect estimate, its standard error estimate, and the warning message for each “Trait” and “Method” combination. For example, if the user wants to analyze two secondary traits, say BMI and BP, using the MLE method (option: “-nsec 2″), there will be 10 columns following the ID column in the output file, and the two main headers will be “BMI_MLE” and “BP_MLE”. If the user wants to analyze these two secondary traits using the LS method (option: “-method LS -nsec 2″), there will be 20 columns following the ID column since there are two versions of the LS method for the secondary trait(s), and the 4 main headers will be “BMI_LS-M”, “BMI_LS-C”, “BP_LS-M”, and “BP_LS-C”. Invalid values are denoted by “NA” in the output file.
The 1st column contains the gene ID and the remaining columns under each main header <Trait_Method> contain the test statistic, the p-value, the genetic effect estimate, its standard error estimate, and the warning message for each “Trait” and “Method” combination. For example, if the user wants to analyze two secondary traits, say BMI and BP, using the MLE method (option: “-nsec 2″), there will be 10 columns following the ID column in the output file, and the two main headers will be “BMI_MLE” and “BP_MLE”. If the user wants to analyze these two secondary traits using the LS method (option: “-method LS -nsec 2″), there will be 20 columns following the ID column since there are two versions of the LS method for the secondary trait(s), and the 4 main headers will be “BMI_LS-M”, “BMI_LS-C”, “BP_LS-M”, and “BP_LS-C”. Invalid values are denoted by “NA” in the output file.
Different types of warning message are as follows:
Singular_covariates: Covariate matrix becomes singular after removing subjects with missing genetic variable or missing “Trait”.
Non-convergence: EM algorithm does not converge.
Low_MAC: MAC is smaller than MAC_LB after removing subjects with missing “Trait”.
Singular_score: The genetic variable contains only one distinct value after removing subjects with missing “Trait”.
Undefined_V: Variance estimate of the score statistic is undefined (numerically 0 or negative).
Pvalue_Fail: p-value calculation fails to obtain the desired level of accuracy.
Var_Null: No valid genetic variable for the test.
- Tests with multiple genetic variables (VT, SKAT, or customized test)The 1st column contains the gene ID and the remaining columns under each main header <Trait_Method> contain the test statistic, the p-value, and the warning message for each “Trait” and “Method” combination.
LOG FILE
Log files include a set of files for each “Trait” and “Method” combination. These files contains the MAC, the score vector, the information matrix, and some other related information. The name of the file is “log_trait_method”: log is the name specified by the option -msglog; method is the method used to analyze the trait. For example, if the user wants to analyze two secondary traits (option “-nsec 2”) under trait dependent sampling (option “-sampling TDS”), say BMI and BP, using the MLE method (option “-method MLE”), there will be 2 log files with names “log_BMI_MLE” and “log_BP_MLE”. If the user wants to analyze these two secondary traits using the LS method (option “-method LS”), then there will be 4 log files with names “log_BMI_LS-M”, “log_BMI_LS-C”, “log_BP_LS-M”, and “log_BP_LS-C”. Invalid values in the output files are denoted by “NA”. The log files for multiples studies can be combined through the accompanying program MASS. We describe below the log file for each test.
- Tests with a single genetic variable (single-variant, T1, T5, or MB test)
The 1st column contains the gene ID and the 2nd column contains the MAC. The remaining columns contain the score statistic and its variance estimate. - VT test
For each gene, “log_trait_method” contains the MAF thresholds, the MACs, the (vector-valued) score statistic, and the information matrix. The following is an example for two genes: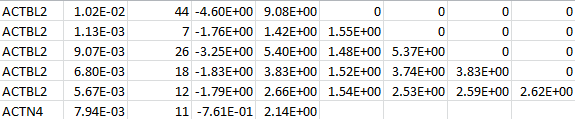
- SKAT test
For a set of SNPs within a gene, “log_trait_method” contains the SNP IDs, the MAFs, the MACs, the number of non-missing genotypes, the counts of homozygous reference, heterozygous, and homozygous alternative, the (vector-valued) score statistic, and the information matrix. The following is an example for two genes: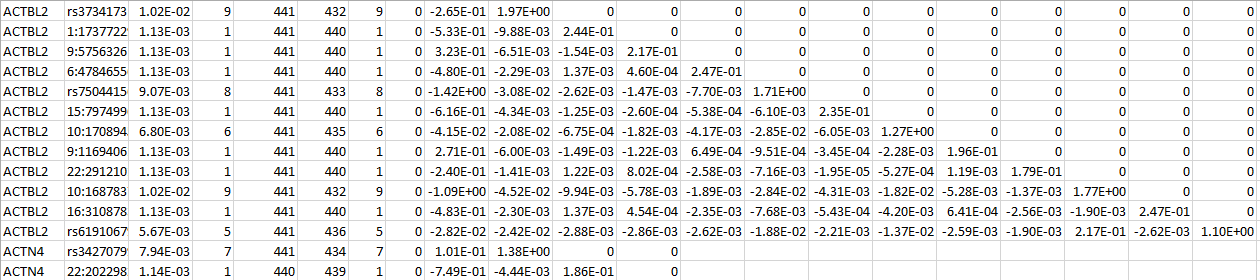
- Customized test
For a set of genetic scores based on the customized weight matrix within a gene, “log_trait_method” contains the weight identifiers, the MACs, the score statistic and the information matrix. The following is an example for two genes: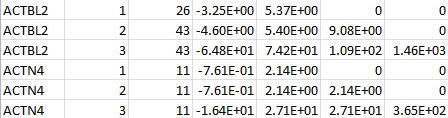
EXAMPLE
The example includes a phenotype file for the sequenced subjects “ldl_seq.txt” , a phenotype file for the non-sequenced subjects “ldl_nonseq.txt” and a genotype file “ldl_geno100.txt“. We want to apply the MLE method to two secondary traits BMI and BP (-nsec 2), apply the MAC lower bound of 2 (-MAC 2) and perform the log-transformation on the primary trait (-log). Enter the command
$ SCORE-SeqTDS -nsec 2 -pfile_seq ldl_seq.txt -pfile_nonseq ldl_nonseq.txt -gfile ldl_geno100.txt -ofile ldl_output.txt -MAC 2 -log
to obtain the results given in the output file “ldl_output.txt” , and the log files “log_BMI_MLE” and “log_BP_MLE” .
Alternatively, we can use “ldl_nonseq2.txt” as the phenotype file for the non-sequenced subjects and obtain the same results by entering the command
$ SCORE-SeqTDS -nsec 2 -pfile_seq ldl_seq.txt -pfile_nonseq ldl_nonseq2.txt -gfile ldl_geno100.txt -ofile ldl_output.txt -MAC 2 -log
MULTIPLE CPUs
If there are a large number of genetic variables, it is preferable to use multiple CPUs. To this end, the user can divide the genotype file into several files, each of which contains a subset of the original genetic variables. Then the software can be run separately for each genotype file on a different CPU. In this way, it takes approximately 1/K (K is the number of jobs that are split into) of the original time to complete the whole set of analysis. We provide a R script to illustrate this approach. In that example, 100 genotype files are created from the original genotype file and 100 jobs are submitted simultaneously to multiple CPUs.
REFERENCE
Dan-Yu Lin, Donglin Zeng, and Zheng-Zheng Tang (2013). Quantitative Trait Analysis in Sequencing Studies Under Trait-Dependent Sampling. Proceedings of the National Academy of Sciences of the United States of America, 110, 12247-12252.
DOWNLOAD
SCORE-SeqTDS for 64-bit x86 based Linux [updated November 18, 2015]
Example files [updated November 27, 2013]
VERSION HISTORY
| Version | Date | Description |
|---|---|---|
| 1 | May 23, 2012 | First version released |
| 2 | Sep 04, 2012 | Added the option “-sampling” to specify the type of sampling design.
Added the option “-rounding” to reduce the number of unique values on the primary trait. Added the variable threshold (VT) test. |
| 3 | Mar 20, 2013 | Added the SKAT test.
Added the customized test. Allowed the covariates of traits to be different. |
| 4 | Nov 27, 2013 | Changed the format of the phenotype file for non-sequenced subjects.
Changed the format of the output and the log files. Changed the name of the option “-burden” to “-test”. |
| 4.1 | Feb 23, 2014 | Added the number of observation for each SNP in the SKAT log file. |
| 5 | June 01, 2015 | Added the options –INT and –R-INT. |
| 7 | July 29, 2015 | Added number of samples (#Samples) in the heading for the all the log files.
In the SNP log output, added columns for counts of homozygous reference, heterozygous, and homozygous alternative. |
| 7.1 | November 18, 2015 | Output statistics for monophonic sites in the SNP log file.
In the SNP log file, calculate MAC under the additive model instead of the dominant model. For the secondary trait analysis, the quality-control statistics are based on the subjects with non-missing secondary trait. Modify the covariance estimator for score statistics when the trait is continuous. |
